Bioinformatic Data Analysis
The GAC GAC assists with the analysis of data from RNA Seq, Exome Seq, Whole Genome Seq, ChiP Seq, ATAC Seq, and other emerging genomic applications.
Examples of projects the GAC has undertaken:
- Single-cell RNA Seq to identify signaling pathways that differentially promote or restrict BKV infection
- Viral diversity and the characterization and discovery of novel viruses
- Custom pipeline development to detect low-frequency drug resistance variants in HIV RT gene in prophylaxis clinical trial samples.
- Variant analysis in samples from pediatric patients with sepsis
- Transcriptional analysis of serum from pediatric and adult influenza vaccine efficacy studies
- Integrative ATAC Seq, ChIP Seq, and RNA Seq analysis of glucocorticoid effects in neural stem cells
- De Novo transcriptomics of opossum kidney cells
Genomics Education
The GAC teaches NGS workshops in collaboration with the CRC.
Current workshop schedules are available.
High Throughput Computing
The GAC collaborates with the CRC and the Pittsburgh Supercomputing Center (PSC) in developing high-throughput computing solutions. Examples of GAC’s role include membership in HTC’s advisory committee and hosting cloud computing workshops such as the NIH STRIDES initiative.
Team Science Projects
The GAC has expertise in the complex requirements of NGS data-centric team science projects from NGS data hosting to development of bioinformatics pipelines, metadata annotation, data harmonization and FAIR principles for data sharing. An example of a cancer-focused team science project - supported by the Cancer Center Support Grant (CCSG) and the Cancer Bioinformatics Services (CBS) - is our role in the Data Coordination Center (DCC) for the multi-site, multi-omics AURORA metastatic breast cancer project (PI: Adrian Lee, Director, IPM)
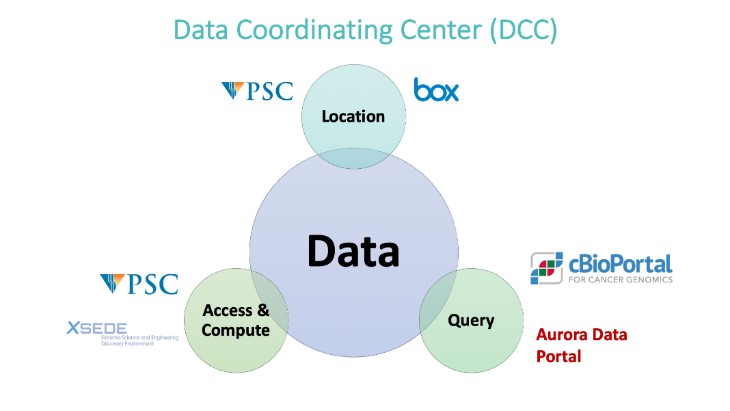
The GAC is seeking similar opportunities to collaborate with Health Science investigators in non-cancer areas such as psychiatry, aging, neurology, cardiology and others.
